Introduction
The rapid escalation of the SARS-CoV-2 virus underscored the urgent need for sophisticated and effective diagnostic tools for COVID-19. The swift dissemination of the virus, coupled with a scarcity of dependable testing methodologies, presented significant hurdles in disease detection, placing immense pressure on healthcare systems globally. The integration of Artificial Intelligence (AI) into medical image analysis has offered a promising avenue to alleviate diagnostic burdens. However, traditional AI approaches frequently necessitate centralized data repositories and model training, leading to increased computational demands and potential privacy vulnerabilities. A key challenge in today’s interconnected world is the secure exchange of sensitive patient data across international healthcare institutions while upholding stringent privacy regulations. Developing collaborative models that prioritize data privacy is paramount for advancing global deep learning initiatives in healthcare.
To navigate these challenges, this paper introduces an innovative framework grounded in blockchain technology and federated learning models. Federated learning addresses computational complexity by enabling decentralized model training, while blockchain ensures distributed data management with robust privacy mechanisms. Specifically, our proposed Federated Learning Ensembled Deep Five Learning Blockchain model (FLED-Block) framework is designed to gather data from diverse medical centers, construct a predictive model utilizing a hybrid capsule learning network, and deliver precise diagnoses while rigorously safeguarding patient privacy and facilitating secure data sharing among authorized entities.
Extensive experimentation using lung CT images demonstrates the superior performance of the FLED-Block model when benchmarked against established models such as VGG-16 and 19, AlexNet, ResNet-50 and 100, Inception V3, DenseNet-121, 119, and 150, MobileNet, and SegCaps. The FLED-Block framework achieved remarkable results in predicting COVID-19, attaining an accuracy of 98.2%, precision of 97.3%, recall of 96.5%, specificity of 33.5%, and an F1-score of 97%. These results highlight its effectiveness in COVID-19 detection while maintaining robust data privacy across heterogeneous user networks.
Keywords: image processing, artificial intelligence, federated learning, blockchain, privacy preservation, capsule learning model, COVID-19 diagnosis
The Imperative for Advanced COVID-19 Diagnostic Models
The COVID-19 pandemic stands as a stark reminder of the devastating impact infectious diseases can have on global populations. The Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) has affected millions, causing widespread health crises and placing unprecedented strain on healthcare infrastructures worldwide. India, for example, has borne a significant burden, ranking among the nations with the highest number of confirmed cases and fatalities. The sheer volume of infections rapidly overwhelmed healthcare providers, highlighting critical shortcomings in diagnostic capabilities and preparedness. Clinicians faced immense difficulties in accurately and promptly identifying COVID-19-positive individuals, primarily due to the limitations of existing testing models.
The diagnosis of COVID-19 is complex, relying on a combination of clinical evaluations, epidemiological history, Computed Tomography (CT) scans, and pathogenic tests. However, CT scans, while valuable, often present overlapping visual features with other pulmonary conditions, complicating differential diagnosis. Furthermore, the symptomatic presentation of COVID-19 is highly variable, and the virus’s rapid transmission necessitates swift and accurate diagnostic processes.
This context underscores the critical need for efficient data sharing among hospitals to enhance diagnostic accuracy. However, the sensitive nature of patient data introduces significant challenges to secure data exchange and the development of robust, globally applicable diagnostic models. Current research is often hampered by the inability to share data effectively and ethically train comprehensive diagnostic AI models. AI-driven diagnostic solutions are further constrained by the difficulties in accessing diverse datasets from multiple sources. The absence of privacy-preserving methodologies within healthcare systems restricts the availability of sensitive patient information, impeding collaborative research and development efforts.
Existing Artificial Intelligence (AI) techniques are being actively explored to address the diagnostic ambiguities inherent in COVID-19 and similar diseases. However, many traditional AI techniques depend on large datasets from single, centralized sources to effectively train deep learning models for accurate predictions. Data homogeneity from single sources can limit the feature distribution variance, potentially leading to higher misclassification rates and impacting diagnostic accuracy. Data sharing across multiple hospitals could mitigate these issues, but stringent security and privacy concerns often prevent healthcare institutions from sharing data, even for critical research purposes. This necessitates advancements in AI methodologies that prioritize collaborative learning while rigorously maintaining data privacy.
The Motivation for Federated Learning and Blockchain Integration
Recent studies from the World Health Organization (WHO) highlight COVID-19 as a primarily pulmonary disease, often resulting in characteristic honeycomb-like lung patterns. Even after recovery, many individuals experience long-term pulmonary impairment. Our research is driven by the critical need to accurately classify COVID-19-related lung patterns, ensuring that experienced radiologists are not overburdened and infections are not overlooked. Furthermore, we aim to facilitate data sharing for training more powerful deep learning models while strictly adhering to data privacy regulations. The development of an automated, deep learning-based model for COVID-19 identification that supports secure data sharing is a primary objective.
We identify several key challenges that motivate our proposed framework:
- Privacy Limitations: Existing privacy protocols often hinder the availability of personal patient information crucial for training robust diagnostic models.
- Federated Model Training: Effectively leveraging blockchain networks to train global diagnostic models in a federated manner is a complex challenge.
- Data Scarcity and Model Improvement: Obtaining sufficient training data to significantly enhance prediction model accuracy, directly impacting diagnostic reliability, remains a major hurdle.
- Pattern Recognition Complexity: Accurately identifying subtle and complex patterns indicative of COVID-19 in lung scans is inherently difficult.
These challenges have spurred the development of our collaborative deep learning model, designed to accurately diagnose COVID-19 cases and facilitate secure result sharing while guaranteeing the privacy and security of participating healthcare institutions. This paper introduces the FLED-Block framework, which enables joint learning from diverse CT image datasets acquired from multiple sources. A novel capsule-deep extreme learning network is integrated to enhance image segmentation and classification. Capsule networks are particularly well-suited for identifying subtle anomalies in medical images. While traditional deep learning models often require vast amounts of data for effective training, capsule networks can improve performance even with smaller datasets by enhancing feature extraction within the model’s internal layers. To further improve diagnostic accuracy, our system synergistically combines the strengths of capsule networks and extreme learning machines (ELMs). ELMs replace conventional dense classification layers with more efficient and accurate mechanisms, leveraging the robust feature maps generated by the capsule network. To address critical privacy concerns, we employ federated learning methodologies to distribute the trained model across a decentralized network, ensuring data privacy at each participating node.
Key Contributions of this Research:
- Ensemble Capsule Network Algorithm: We propose a novel algorithm that leverages an ensemble of capsule networks to detect and classify COVID-19 patterns in CT images from multiple sources. The integration of extreme learning machines within the capsule network enhances feature extraction, leading to improved classification accuracy.
- Blockchain-Powered Federated Data Sharing: This paper introduces a blockchain-enabled data collection and sharing unit. This unit securely gathers data from diverse, heterogeneous sources and employs federated learning to maintain data privacy among organizations while enabling high-accuracy global model training.
- Empirical Validation of Superior Performance: The effectiveness of the proposed algorithm is rigorously validated through extensive experimentation using datasets from various sources. Performance metrics are comprehensively evaluated and compared against existing deep learning algorithms, demonstrating the superior performance of our approach.
This research article presents a blockchain-empowered federated framework designed to improve the recognition of heterogeneous CT images from multiple sources and facilitate secure data sharing among healthcare institutions while maintaining stringent privacy and security protocols. The integration of ensembled capsule networks and ELMs optimizes feature extraction and classification, significantly enhancing the detection of COVID-19 across diverse, publicly available CT image datasets.
The remainder of this paper is structured as follows: Section 2 reviews related work in decentralized networks and federated learning. Section 3 details the FLED-Block model, including data normalization techniques, the ensembled learning model architecture, and the blockchain-based federated data sharing mechanism. Section 4 presents and analyzes the experimental results, findings, and comparative performance evaluations. Finally, Section 5 concludes the paper, summarizing the results and outlining directions for future research and enhancements.
Related Works: Decentralized Networks and Federated Learning in Healthcare
Supriya et al. (2021) explored contemporary medical imaging analysis techniques, emphasizing their role in prediction, e-treatment, stage classification, remote monitoring, and secure data transmission. Their study highlighted the increasing utilization of supervised learning classifiers such as Support Vector Machines (SVM), Decision Trees (DT), K-Nearest Neighbors (KNN), and Artificial Neural Networks (ANN) in medical imaging. Furthermore, they underscored the critical role of blockchain technology in facilitating public access to medical data and secure data transfer globally through distributed networks. The authors concluded that these technological advancements are pivotal in modern medical image transmission and are transforming healthcare service delivery.
Recognizing the limitations in disseminating crucial COVID-19 management and prevention information due to patient privacy concerns, Tripathi et al. (2021) proposed a privacy-centric architecture based on federated learning and blockchain. Their framework aimed to enhance public communication and provide alternative channels for distributing COVID-19-related information. The proposed architecture effectively addresses the challenge of data silos and enables the development of shared models while respecting data ownership and privacy. Their findings indicated that this infrastructure is robust against information security and privacy breaches, offering a secure and efficient solution for sensitive health data management.
Georgios et al. (2021) developed PriMIA, an open-source software framework based on federated learning specifically for medical imaging. PriMIA is designed to handle multi-source pediatric radiology data for classification purposes. Their framework incorporates a deep convolutional neural network (DCNN) trained to classify different stages of cardiac disease using a pediatric chest X-ray image database. The DCNN is trained using a gradient-based model to detect chest diseases at early stages, showcasing the potential of federated learning in pediatric diagnostics.
Shichang et al. (2021) introduced a blockchain-based security model for detecting malicious nodes in federated learning systems. Their model integrates competing vote authentication techniques and aggregation strategies to enhance the security of federated learning processes. A key feature of this model is the optimization of communication costs associated with transferring large datasets during federated learning, effectively mitigating common attacks such as “free-riding attacks” and “model poisoning attacks,” thus enhancing the reliability and security of federated learning in distributed environments.
Kim et al. (2020) proposed a “Blockchain Federated Learning” (BlockFL) architecture for decentralized federated learning. BlockFL is designed to overcome the single point of failure vulnerability inherent in traditional federated learning systems by decentralizing the federated scope. The architecture incorporates local training outcomes into a verification procedure to establish trust among devices in public networks, enhancing the robustness and trustworthiness of federated learning in open environments.
Bao et al. (2020) presented FLchain, a centralized, publicly auditable, and robust federated learning ecosystem. FLchain replaces the traditional centralized coordinator in federated learning with blockchain technology, introducing decentralization and transparency. This approach leverages blockchain to enhance the integrity and auditability of federated learning processes, fostering greater trust and accountability in collaborative model training.
Majeed and Seon (2019) introduced a channel-specific ledger-based blockchain architecture for federated learning, enabling global model learning through blockchain channels. In this architecture, each local parameter of the model is stored as blocks in a channel-specific ledger, adhering to decentralized data management principles. This approach allows for secure and organized management of model parameters across distributed nodes in a federated learning network.
Martinez et al. (2020) explored the use of cryptocurrency and federated learning to address data privacy and security challenges in collaborative machine learning. The authors proposed a detailed methodology incorporating an off-chain record database for scalable gradient recording and reward mechanisms. This system leverages cryptocurrency to incentivize participation and ensure fairness in federated learning, while maintaining data privacy and security through decentralized model training.
Abdul Salam et al. (2021) developed a novel federated learning algorithm specifically for COVID-19 patients. Their neural network is pre-trained using chest X-ray (CXR) images from patients with abnormalities to predict mortality risk and facilitate e-treatment strategies. However, the authors noted that the predictor’s limitation is its reduced efficacy with very large datasets, highlighting a potential area for improvement in scalability.
Parnian et al. (2020) presented COVID-CAPS, a model based on capsule networks, designed to overcome the limitations of CNN-based models when dealing with smaller datasets. COVID-CAPS is specifically tailored for detecting COVID-19-positive cases from X-ray images. Their research demonstrated that the COVID-CAPS model outperforms conventional networks, particularly when model parameters are carefully adjusted for optimal performance in medical image analysis.
He et al. (2020) conducted an experimental study on automated federated learning (AutoFL) using neural architecture search (NAS) algorithms and federated NAS (FedNAS) algorithms. AutoFL aims to enhance the quality and efficiency of local machine learning models that collaboratively update their model parameters. Their findings indicated that default parameters for local machine learning models are often suboptimal in a federated context, particularly for non-IID (non-independent and identically distributed) client data, emphasizing the need for tailored optimization strategies in federated learning environments.
FLED-Block Model: Federated Learning and Blockchain for COVID-19 Diagnosis
Healthcare organizations and hospitals are understandably cautious about sharing patient data due to stringent privacy regulations and ethical considerations. However, the development of effective deep learning models for disease diagnosis necessitates access to large and diverse datasets. Processing vast amounts of data can lead to increased computational complexity and potential performance bottlenecks. To address these challenges, our research proposes the FLED-Block model, a novel framework designed for training and sharing global diagnostic models in a privacy-preserving and computationally efficient manner. Figure 1 illustrates the architecture of the proposed FLED-Block framework.
Figure 1. Proposed Framework of the FLED-Block.
Alt Text: FLED-Block framework architecture for COVID-19 diagnosis using federated learning and blockchain, illustrating data flow from hospitals through normalization, ensembled capsule network training, ELM classification, and secure model sharing via blockchain.
The FLED-Block architecture facilitates the collaborative collection of data from various hospitals, accommodating the inherent variability in CT scanner types and imaging protocols. The initial step involves rigorous data normalization, employing both spatial normalization and signal normalization techniques to standardize the diverse CT image datasets. Subsequently, advanced deep learning algorithms are utilized to identify COVID-19 patterns within lung CT images. For enhanced generalization and robust feature extraction, image segmentation and training are performed using an ensembled capsule network, which has demonstrated superior performance compared to traditional learning models.
The segmented COVID-19 images are then classified using extreme learning machines (ELMs). ELMs, known for their single hidden layer architecture and self-tuning capabilities, offer efficient and accurate classification. Finally, to address the critical issue of data privacy and facilitate collaborative model development, we integrate federated learning techniques. This approach enables the construction of a global model by aggregating locally trained models from participating hospitals, without requiring direct access to sensitive patient data. Federated learning allows hospitals to maintain the privacy of patient information while securely exchanging model weights and gradients via blockchain technology. This decentralized architecture ensures secure and private data communication among hospitals, safeguarding patient confidentiality while fostering collaborative AI model development.
In summary, the proposed FLED-Block framework operates through three key stages: (1) secure data collection from diverse medical healthcare centers; (2) hybrid capsule learning network-based model training for segmentation and classification of COVID-19 images; and (3) collaborative sharing of the hybrid model using blockchain with federated learning, ensuring stringent privacy preservation for all participating organizations.
Materials and Methodologies: Datasets and Normalization
Artificial intelligence is playing an increasingly critical role in clinical diagnostics, particularly in image-based disease detection. Deep learning algorithms, in particular, require substantial amounts of data for effective training and validation. To rigorously evaluate the proposed FLED-Block model, we utilized diverse heterogeneous CT image datasets. The following sections detail the characteristics of Datasets 1, 2, and 3, which were employed in our study.
Dataset-1
The first dataset comprises 34,006 CT scan slices collected from three hospitals, representing 89 individuals. Of these slices, 28,395 are from patients confirmed to be COVID-19-positive. The data were acquired using six different CT scanners, encompassing scans from 89 unique patients. Among these patients, 68 tested positive for COVID-19, while 21 tested negative. Figure 2 visually represents sample CT images from Dataset-1.
Figure 2. Computed Tomography Specimen Images—Dataset-1.
Alt Text: Sample CT scan images from Dataset-1, exhibiting varying lung conditions and image characteristics from different scanners, used for COVID-19 detection model training and validation.
Dataset-2
Dataset-2 consists of chest CT scans from patients with confirmed COVID-19 infections. These unenhanced CT images were collected from patients with positive Reverse Transcription Polymerase Chain Reaction (RT-PCR) tests and accompanying clinical symptoms between March 2020 and January 2021. Common pre-existing conditions among these patients included hypertension, coronary heart disease, diabetes, and interstitial pneumonia or emphysema. CT examinations were performed in “Helical” mode using a NeuViz 16-slice CT scanner (Neu soft medical systems), and all images are in DICOM format, comprising 512 x 512 pixel 16-bit grayscale images.
Dataset-3
Dataset-3 includes 349 COVID-19 CT scans from 216 patients and 463 non-COVID-19 CT images. These datasets represent initial images acquired at the point of care during an epidemic situation from patients with RT-PCR confirmed SARS-CoV-2 infection. Datasets are further detailed in references 46 and 47.
Given the heterogeneity of data sources, we implemented a normalization approach to standardize CT scan images for effective federated learning. Table 1 provides an overview of the datasets used to evaluate the proposed federated learning model. Figure 3 displays sample images from Datasets 2 and 3.
Table 1. Summary of the Dataset Details Used for the Proposed Research.
| Datasets | Source of datasets | No of COVID-19 patients | No of Non-COVID-19 infections | Image formats | Number of patients | Total number of CT scan images |
|---|---|---|---|---|---|---|
| Datasets-1 | CC-19 datasets [ ] | 63 | 26 | CT Scan images | 89 | 34,006 |
| Datasets-2 | COVID-19-CT datasets | 783 | 217 | DICOM Images | 1,000 | 45,002 |
| Dataset-3 | COVID-19 CT datasets [ ] | 216 | 463 | CT Scan Images/DICOM | 689 | 3,490 |
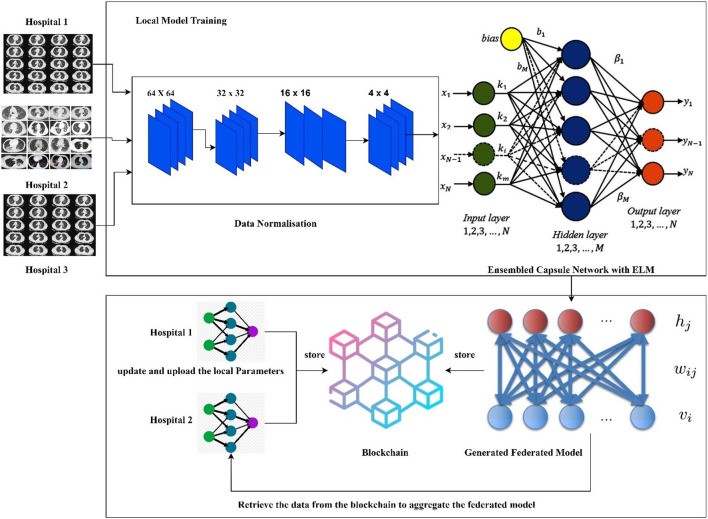
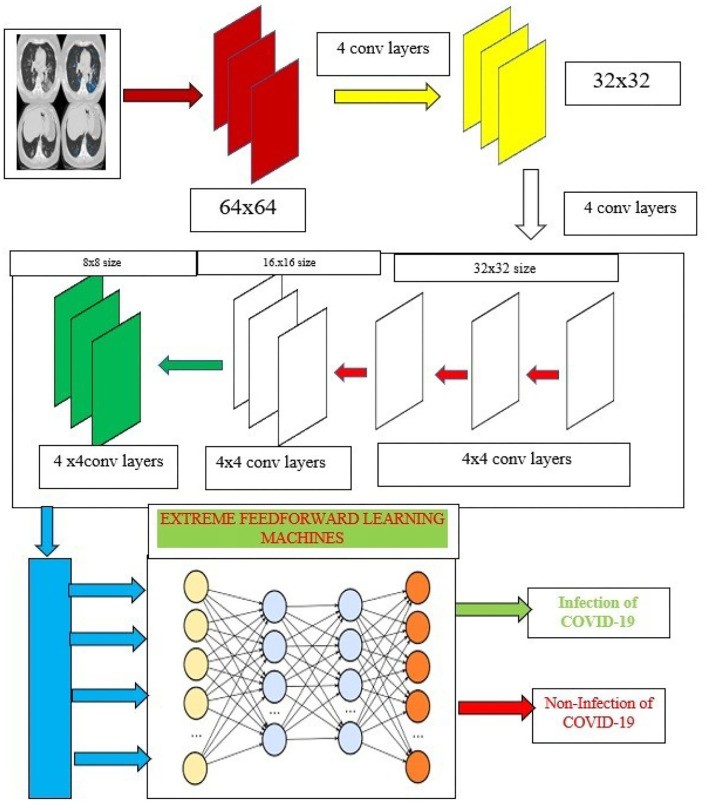
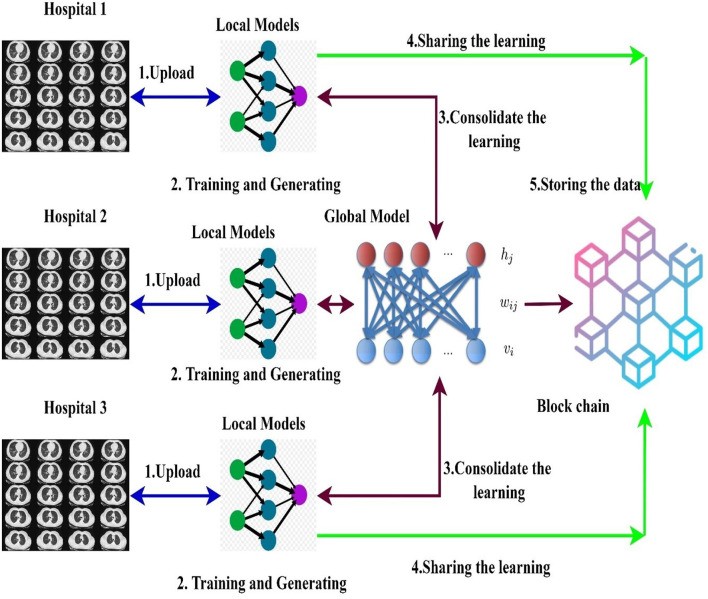
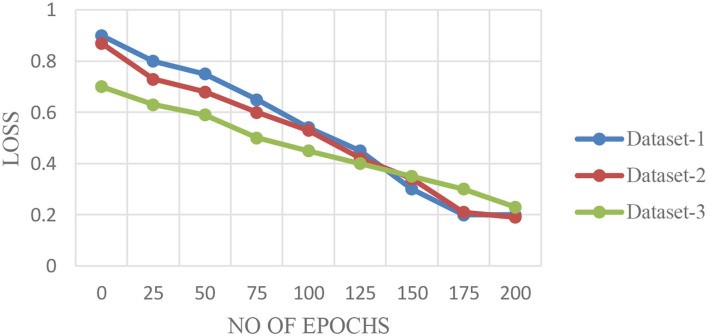
Figure 3. Computed Tomography Specimen Images—Datasets 2 & 3.
Alt Text: Representative CT scan images from Datasets 2 and 3, showcasing the diversity in image quality, patient conditions, and data formats used for training the federated COVID-19 detection model.
Data Normalization Techniques
We adopted the data normalization techniques described by Krizhevsky et al. (2012) to address the challenges posed by heterogeneous data sources. Effective normalization is crucial for enhancing the performance of federated learning models, particularly when dealing with diverse medical image datasets. Following Krizhevsky et al., we employed two primary normalization techniques: signal normalization and spatial normalization.
Signal Normalization Technique
Signal normalization adjusts voxel intensity values based on the lung window, a standard practice in CT image analysis. CT scans are typically represented in Hounsfield Units (HU), and lung windowing utilizes window level (WL) and window width (WW) parameters to optimize visualization of lung tissues. We normalized image intensity values using Equation 1:
| $O_{normalized} = (O – WL) / WW$ | (1) |
Where $O_{normalized}$ represents the normalized image intensity, and $O$ is the original image intensity. For our experiments, we selected a lower bound window size within the range of [-0.05, 0.5], optimizing the contrast and visibility of lung features relevant to COVID-19 diagnosis.
Spatial Normalization Technique
Spatial normalization addresses variations in image dimensions and resolutions across different CT scans. We standardized all CT scan images to a consistent resolution of 332 × 332 × 512 mm³, as suggested by Krizhevsky et al. (2012). This spatial normalization ensures that all datasets, regardless of their original format, are transformed into a uniform format suitable for federated learning. Standardizing image dimensions improves model learning efficiency and overall performance by eliminating variability arising from differing image sizes and resolutions.
Ensembled Capsule-Based Model Training: Enhancing Feature Extraction and Classification
Deep learning frameworks have gained immense popularity in image analysis due to their powerful feature extraction and classification capabilities. Convolutional Neural Networks (CNNs) have been particularly dominant in image classification tasks. However, CNNs can suffer from limitations due to pooling layers, which may not effectively capture spatial relationships between image features. This can lead to increased computational complexity and potentially impact classifier performance. To overcome these limitations and achieve improved classification accuracy and diagnostic precision, our proposed system integrates the robust feature extraction capabilities of capsule networks with the efficient classification mechanisms of extreme learning machines (ELMs). In this approach, capsule networks are employed to extract detailed and spatially coherent feature maps, while ELMs replace traditional dense classification layers, providing a more efficient and effective approach for COVID-19 prediction.
Capsule Networks: Capturing Spatial Hierarchies in Medical Images
Capsule networks, as introduced by Hinton et al. (2018), address some of the limitations of CNNs by preserving spatial relationships between features. A typical capsule network architecture includes: (1) a convolutional layer for initial feature extraction, (2) a hidden layer for non-linear transformation, (3) a PrimaryCaps layer to form initial capsules representing object parts, and (4) a DigitCaps layer to aggregate information and perform final classification. Figure 4 illustrates the architecture of our proposed training model, incorporating capsule networks and ELMs. The normalized CT images serve as input to our capsule network-based model. The capsule network operates in two primary phases:
Figure 4. Capsule Ensembled ELM Layers for Achieving Feature Extraction and Classification Accuracy.
Alt Text: Architecture of the proposed ensembled capsule network with ELM, showing normalized CT image input, capsule network layers for feature extraction, ELM classification layer, and COVID-19 diagnosis output, highlighting the integration for enhanced accuracy.
Capsule networks represent features as “capsules,” which are groups of neurons whose activity vectors represent the instantiation parameters of a specific type of entity, such as an object or object part. These instantiation parameters can include properties like pose, deformation, texture, and velocity. Crucially, capsule networks encode two key aspects of feature representation:
- Probability of Entity Existence: Capsules represent the likelihood that a particular entity is present in the image.
- Entity Instantiation Parameters: Capsules encapsulate the properties or characteristics of the detected entity.
To encode the spatial relationships between low-level and high-level features within the image, we calculate the weighted input vectors. Equation 2 describes the relationship between input vectors “s,” weight matrix “W,” and component vector “U”:
| $Y{(i,j)} = W{(i,j)} U_{(i,j)} * S_j$ | (2) |
The current capsule “D” is determined by summing the weighted input vectors, as shown in Equation 3:
| $S{(j)} = sum{j} Y{(i,j)} * D{(j)}$ | (3) |
Finally, non-linearity is applied using a squash function, described by Equation 4:
| $Y{(i,j)} = W{i,j} U_{(i,j)} * S_j$ | (4) |
The distribution of information from low-level capsules to high-level capsules is iteratively adjusted based on the output, refining the representation until an optimal distribution is achieved. This dynamic routing mechanism allows capsule networks to effectively learn and represent hierarchical spatial relationships, which are crucial for accurate medical image analysis.
Extreme Learning Machines (ELMs): Efficient and Rapid Classification
To complement the robust feature extraction of capsule networks, we integrate extreme learning machines (ELMs) for image classification. ELMs are single-hidden layer feedforward neural networks known for their efficiency and speed. They operate on the principle of random initialization of input weights and biases, and analytical determination of output weights, eliminating the need for iterative tuning of hidden layer parameters. Figure 4 illustrates the integration of ELMs within our proposed architecture.
ELMs offer significant advantages over other learning models, including support vector machines (SVMs), Bayesian classifiers (BC), K-nearest neighbors (KNN), and random forests (RF), particularly in terms of computational speed and reduced overhead. ELMs achieve high performance with minimal training error and excellent approximation capabilities due to their autotuning properties of weight biases and the use of non-zero activation functions. Detailed descriptions of the ELM working mechanism can be found in Huang et al. (2006) and Wang et al. (2018).
Mathematically, the characteristic of an ELM with L hidden nodes can be represented by Equation 5:
| $fL(x) = sum{i=1}^{L} beta_i h_i(x) = h(x) beta$ | (5) |
Where:
- $x$ is the input vector.
- $beta$ is the output weight vector, defined as: $beta = [beta_1, beta_2, dots, beta_L]^T$ (6)
- $h(x)$ is the output of the hidden layer, given by: $h(x) = [h_1(x), h_2(x), dots, h_L(x)]$ (7)
The output of the ELM, $f_L(x)$, can be calculated using Equation 8:
| $f_L(x) = h(x) beta = h(x) H^T ( frac{1}{C} I + HH^T )^{-1} O$ | (8) |
Where:
- $O$ represents the output target vectors.
- $H$ is the hidden layer output matrix.
- $C$ is a regularization parameter.
The infectious impact of COVID-19 on lungs is effectively classified based on Equation (7). Algorithm 1 presents the pseudo-code outlining the working mechanism of our proposed ensembled network.
Algorithm 1. Pseudo Code for the Proposed Ensembled Algorithm.
| 1 Inputs: Normalized Input Images: I |
|---|
| 2 Output: Presence of COVID-19 diseases on Lungs |
| 3 For n = 0 to Max_iterations n = No of iterations |
| 4 Features F = Capsule(I) // Using Equations (2–4) |
| 5 Output Function = ELM(F) // Using Equation 7 |
| 6 If Output == threshold // User-based threshold |
| 7 COVID-19 is detected |
| 8 Else |
| 9 Normal Condition is detected |
| 10 End |
| 11 End |
| 12 End |
Federated Learning for Global Model Training: Privacy-Preserving Collaboration
This section details the decentralized data sharing mechanism utilizing federated learning across multiple hospitals. Our proposed model leverages federated learning to enable model sharing among hospitals without compromising patient privacy. It also aggregates locally trained models from different healthcare institutions to create a robust global diagnostic model. We consider a scenario with H hospitals and a total dataset d. In our federated model, the ensembled learning model (capsule network and ELM) serves as the global model M. Initially, the weights W of the ELM are randomly distributed to participating hospitals. Figure 5 illustrates the federated learning model employed in our research.
Figure 5. Blockchain Empowered Federated Learning Models Used in the Proposed Framework.
Alt Text: Blockchain-integrated federated learning model architecture, showing data localization at hospitals, local model training, weight aggregation via blockchain, global model update, and secure distribution, emphasizing privacy and collaboration.
Our blockchain-based federated learning framework facilitates collaborative training and sharing of diagnostic models. Each hospital utilizes a global or collaborative model, achieved through federated learning, to integrate the weights of locally trained models. Data is initially collected from multiple sources and processed locally at each hospital, using the normalization techniques described earlier to handle the heterogeneity of CT scan data. After normalization, the ensembled capsule network is used to segment images and train a local model to detect COVID-19 indicators. Crucially, only the weights of these locally trained models are shared across the blockchain network to train a global model. This ensures that sensitive patient data remains within each hospital’s secure environment, upholding privacy regulations.
Mathematically, let H be the number of hospitals, and d be the total dataset, partitioned into training and testing sets as defined in Equations 9 and 10:
| $D{i}^{train} = {(X{i,j}^{train}, Y{i,j}^{train})} text{ where } j=1 text{ to } N{text{train data}}$ | (9) |
|---|---|
| $D{i}^{test} = {(X{i,j}^{test}, Y{i,j}^{test})} text{ where } j=1 text{ to } N{text{test data}}$ | (10) |
The total dataset used for training the global model is the union of training and testing datasets from all hospitals, as shown in Equation 11:
| $D{(i)} = D{i}^{train} cup D_{i}^{test}$ | (11) |
Since data $D_{(i)}$ is collected from heterogeneous sources, the data distribution across hospitals may be unequal. In each communication round of federated learning, the weights W of the ELM are distributed to participating hospitals. Hospitals train local models using these weights and store model updates on the blockchain network. Weights are iteratively updated and recorded on the blockchain for each round of federated learning. The weight update process is mathematically represented by Equation 12:
| $eta = W_i – W_l$ | (12) |
Where $W_i$ represents the distributed weights of the global model, and $W_l$ represents the weights of the local model. Finally, all local models stored on the blockchain are aggregated to form a new, improved global learning model, leveraging the principles of ELM and implemented within our proposed deep learning algorithm.
Blockchain Framework for Secure Federated Learning
The proposed framework incorporates blockchain architecture to provide a secure and efficient data retrieval and sharing process for federated learning. This enables multiple hospitals to collaboratively train diagnostic models, enhancing disease detection capabilities. Building upon multi-organization blockchain architectures, our approach focuses on secure data retrieval and sharing within the federated learning context.
Blockchain-Based Data Retrieval Process
Each hospital contributes data (in the form of local model updates) and stores it as a transaction on the blockchain network. Data retrieval from blockchain nodes depends on two key parameters: the distance between nodes (d) and the hospital ID (ID). Based on the geographical or network distance between hospitals, a unique ID is generated and managed by the blockchain. The blockchain maintains log tables to store the unique IDs of participating hospitals. Data is retrieved from neighboring hospitals identified by their unique IDs, facilitating efficient and localized data access.
Mathematically, hospitals are represented as nodes X, partitioned into different communities. The distance between nodes, representing hospitals, is calculated using Equation 13:
| $d(X{(i)}, X{(j)}) = frac{sum{p,q, in {X{(i)} cup X{(j)} – X{(i)} cap X{(j)}}} text{Attributes of the Nodes}}{sum{p,q, in {X{(i)} cup X{(j)}}} text{Attributes of Nodes} * log(X{(i)}, X{(j)})}$ | (13) |
Where $X{(i)}$ and $X{(j)}$ represent neighboring hospitals located at positions i and j, differentiated by their unique IDs.
In the next phase, a consensus process is employed to train the FLED-Block model using the retrieved local models. This collaborative training process involves all participating nodes working together to refine the global model. The blockchain consensus mechanism provides proof of work, ensuring data integrity and secure sharing across the network. To verify the quality of local models during collaborative training, we calculate the Mean Prediction Accuracy Error (MPAE). MPAE serves as a metric for assessing the predictive performance of the models. To ensure data confidentiality, all data stored on the blockchain nodes is encrypted using high-random chaotic public and private keys. All transactions are subject to MPAE validation, and validated transactions are recorded in the distributed ledger of the blockchain, maintaining transparency and auditability.
Blockchain-Based Data Sharing Process
Security is paramount in the data sharing process between requesting and source hospitals. Instead of sharing raw patient data, hospitals share only the learned models with authorized requesters, preserving patient privacy. Hospitals communicate through the blockchain network, and a consensus algorithm facilitates federated learning from distributed data. Data providers’ and requesters’ information is securely stored in the blockchain nodes. To further enhance data privacy, only learning models (model weights and updates) are shared, not original patient data. In the initial phase, each hospital uploads image datasets for collaborative learning. In the subsequent phase, hospitals share their locally trained model weights with the blockchain. Federated learning is then used to aggregate these local models into a robust global model, ensuring collaborative model improvement without direct data exchange.
Experimental Results: Validating FLED-Block Performance
The proposed FLED-Block model was developed using the open-source TensorFlow Federated version 2.1.0. Classical deep learning models used for comparison were implemented using TensorFlow version 1.8, with Keras as the backend for both sets of models. All experiments were conducted on a high-performance PC workstation equipped with an Intel Xeon CPU, NVIDIA Titan GPU, 16GB RAM, and a 3.5 GHz operating frequency. Each dataset was partitioned into 70% for training, 20% for testing, and 10% for validation. The 70% training split from each of the three datasets (Dataset 1: 23,804 images, Dataset 2: 31,501 images, Dataset 3: 2,443 images) was used to train both the proposed FLED-Block model and the classical models. The FLED-Block model was implemented on a federated blockchain environment using Python 3.9.1. The blockchain user interface was developed using CSS. This experimental setup was used for algorithm validation and testing. The performance of the FLED-Block architecture was evaluated using standard performance metrics, including accuracy, precision, recall, specificity, and F1-score. These metrics quantify the model’s ability to correctly differentiate between COVID-19 and non-COVID infections. Table 2 presents the mathematical expressions used for calculating these performance metrics.
Table 2. Comparative Analysis of the Different Algorithms in Detecting COVID-19 Using Dataset 1.
| Algorithm | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|
| VGG-16 | 0.8269 | 0.833 | 0.8234 | 0.170 | 0.832 |
| VGG-19 | 0.833 | 0.843 | 0.823 | 0.173 | 0.840 |
| AlexNet | 0.834 | 0.823 | 0.814 | 0.189 | 0.826 |
| ResNet-50 | 0.845 | 0.823 | 0.832 | 0.164 | 0.834 |
| ResNet-100 | 0.849 | 0.843 | 0.834 | 0.167 | 0.838 |
| Inception V3 | 0.80 | 0.82 | 0.821 | 0.190 | 0.801 |
| DenseNet-121 | 0.82 | 0.83 | 0.834 | 0.167 | 0.812 |
| DenseNet-119 | 0.78 | 0.793 | 0.80 | 0.200 | 0.80 |
| DenseNet-150 | 0.81 | 0.802 | 0.794 | 0.80 | 0.73 |
| MobileNet | 0.782 | 0.784 | 0.778 | 0.783 | 0.778 |
| SegCaps | 0.89 | 0.934 | 0.923 | 0.07 | 0.930 |
| Proposed Model (FLED-Block) | 0.982 | 0.973 | 0.965 | 0.0335 | 0.970 |
In medical diagnostic systems, high accuracy, precision, and recall are essential. To mitigate overfitting and improve model generalization, we implemented an early stopping method. This technique terminates model training when validation performance shows no improvement for a predefined number of consecutive epochs, preventing the model from memorizing training data and enhancing its ability to generalize to unseen data.
Results and Findings: Superior Performance of FLED-Block
The performance of the proposed FLED-Block architecture was validated in three key aspects: (1) evaluation of performance metrics across different CT datasets; (2) analysis of loss validation curves (LVCs) to assess model training behavior; and (3) comparative performance analysis against existing deep learning algorithms. This section presents the performance results of the FLED-Block model using Datasets 1, 2, and 3, as illustrated in Figures 6A–C.
Figure 6. (A–C) Training—Validation Curves for the Proposed Algorithm for Different Datasets.
Alt Text: Training and validation accuracy curves for FLED-Block model on Datasets 1, 2, and 3, showing high accuracy and minimal overfitting across datasets, demonstrating robust learning.
Figures 6A–C display the training and validation accuracy curves for the FLED-Block model across the three datasets. Figure 7 shows the corresponding loss validation curves. From Figure 6A (Dataset 1), the Root Mean Square Error (RMSE) between the training and validation models is approximately 0.001, indicating excellent model convergence and minimal overfitting. Similar characteristics are observed in Figure 6C (Dataset 3). Dataset 2, being larger, exhibits a slightly higher RMSE of 0.0014 in Figure 6B, but still demonstrates robust convergence. The average diagnostic accuracy of the FLED-Block model was consistently high across all datasets: 98.5% for Dataset 1, 98.3% for Dataset 2, and 98.5% for Dataset 3.
Figure 7. Loss Validation Curves for the Proposed Algorithm for Different Datasets.
Alt Text: Loss validation curves for FLED-Block model on Datasets 1, 2, and 3, showing low and stable validation loss across datasets, indicating effective model training and generalization.
The robust performance of the FLED-Block model across multi-source heterogeneous datasets can be attributed to the effective feature extraction capabilities of capsule networks and the efficient classification provided by ELMs. Figure 8 summarizes the key performance metrics of the FLED-Block model across the three datasets. The average performance metrics consistently demonstrate high accuracy (98.4% to 98.5%), precision (97% to 98%), and recall (97.5% to 98%). However, the model exhibits relatively lower specificity and higher false alarm rates across all three datasets, indicating a potential area for further refinement to reduce false positives.
Figure 8. Performance Metrics of the Proposed Algorithm with the Different Datasets.
Alt Text: Bar chart comparing accuracy, precision, recall, specificity, and F1-score of FLED-Block model across Datasets 1, 2, and 3, highlighting consistently high performance in accuracy, precision, recall, and F1-score.
To rigorously validate the superiority of the FLED-Block model, we conducted a comprehensive comparative analysis against established deep learning algorithms, including VGG-16/VGG-19, AlexNet, DenseNet-121/119/150, ResNet-50/100, and SegCaps. Table 2 presents the performance comparison for Dataset 1. The FLED-Block model significantly outperformed all other models, achieving the highest performance metrics, surpassing even SegCaps and ResNet-100, which also demonstrated strong performance. Table 3 and Table 4 present comparative results for Datasets 2 and 3, respectively, showing similar trends.
Table 3. Comparative Analysis of the Different Algorithms in Detecting COVID-19 Using Dataset 2.
| Algorithm | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|
| VGG-16 | 0.797 | 0.783 | 0.7563 | 0.289 | 0.789 |
| VGG-19 | 0.732 | 0.743 | 0.723 | 0.273 | 0.7390 |
| AlexNet | 0.804 | 0.783 | 0.784 | 0.229 | 0.806 |
| ResNet-50 | 0.80 | 0.801 | 0.802 | 0.200 | 0.812 |
| ResNet-100 | 0.840 | 0.838 | 0.836 | 0.177 | 0.82 |
| Inception V3 | 0.678 | 0.677 | 0.675 | 0.675 | 0.681 |
| DenseNet-121 | 0.790 | 0.784 | 0.779 | 0.221 | 0.79 |
| DenseNet-119 | 0.777 | 0.781 | 0.78 | 0.229 | 0.774 |
| DenseNet-150 | 0.80 | 0.792 | 0.789 | 0.728 | 0.73 |
| MobileNet | 0.782 | 0.784 | 0.783 | 0.773 | 0.753 |
| SegCaps | 0.87 | 0.92 | 0.910 | 0.09 | 0.910 |
| Proposed Model (FLED-Block) | 0.982 | 0.973 | 0.965 | 0.335 | 0.970 |
Table 4. Comparative Analysis of the Different Algorithms in Detecting COVID-19 Using Dataset 3.
| Algorithm | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|
| VGG-16 | 0.8269 | 0.833 | 0.8234 | 0.170 | 0.832 |
| VGG-19 | 0.833 | 0.843 | 0.823 | 0.173 | 0.840 |
| AlexNet | 0.834 | 0.823 | 0.814 | 0.189 | 0.826 |
| ResNet-50 | 0.845 | 0.823 | 0.832 | 0.164 | 0.834 |
| ResNet-100 | 0.849 | 0.843 | 0.834 | 0.167 | 0.838 |
| Inception V3 | 0.80 | 0.82 | 0.821 | 0.190 | 0.801 |
| DenseNet-121 | 0.82 | 0.83 | 0.834 | 0.167 | 0.812 |
| DenseNet-119 | 0.78 | 0.793 | 0.80 | 0.200 | 0.80 |
| DenseNet-150 | 0.81 | 0.802 | 0.794 | 0.80 | 0.73 |
| MobileNet | 0.782 | 0.784 | 0.778 | 0.783 | 0.778 |
| SegCaps | 0.89 | 0.934 | 0.923 | 0.07 | 0.930 |
| Proposed Model (FLED-Block) | 0.982 | 0.973 | 0.965 | 0.335 | 0.970 |
Across all datasets, the FLED-Block model consistently demonstrated superior performance. While the performance of other deep learning models slightly decreased with larger datasets (Dataset 2), SegCaps, ResNet-100, and particularly the FLED-Block model maintained robust performance, with FLED-Block consistently outperforming all benchmarks. The ensemble of capsule networks and extreme learning machines in the FLED-Block model delivers exceptional accuracy in detecting COVID-19 from multi-source datasets, demonstrating its superiority over other algorithms.
Finally, we compared the FLED-Block model with other blockchain-based learning models in terms of detection accuracy, trust level, and data sharing capabilities. Tables 5–7 present comparative analyses for Datasets 1, 2, and 3.
Table 5. Comparative Analysis Between Blockchain-Based Learning Models for COVID-19 Detection of Diseases Using Dataset 1.
| References | Proposed model in blockchain | Number of cases | Average accuracy performance % | Trust level | Sharing and retrieval |
|---|---|---|---|---|---|
| Parnian et al. (2020) | ResNETS | High | 89.5 | No | No |
| He et al. (2020) | 2D-CNN | High | 85.4 | No | No |
| Rahimzadeh et al. (2021) | Federated Capsule network learning | High | 91 | Medium | Yes |
| Ours (FLED-Block) | Federated Ensembled capsule networks | High | 98.5 | High | Yes |
Table 6. Comparative Analysis Between Blockchain-Based Learning Models for COVID-19 Detection of Diseases Using Dataset 2.
| References | Proposed model in blockchain | Number of cases | Average accuracy performance % | Trust level | Sharing and retrieval |
|---|---|---|---|---|---|
| Parnian et al. (2020) | ResNETS | Very High | 88.4 | No | No |
| He et al. (2020) | 2D-CNN | Very High | 83.3 | No | No |
| Rahimzadeh et al. (2021) | Federated Capsule network learning | Very High | 89 | Medium | Yes |
| Ours (FLED-Block) | Federated Ensembled capsule networks | Very High | 98.5 | High | Yes |
Table 7. Comparative Analysis Between Blockchain-Based Learning Models for COVID-19 Detection of Diseases Using Dataset 3.
| References | Proposed model in blockchain | Number of cases | Average accuracy performance % | Trust level | Sharing and retrieval |
|---|---|---|---|---|---|
| Parnian et al. (2020) | ResNETS | High | 89.5 | No | No |
| He et al. (2020) | 2D-CNN | High | 85.4 | No | No |
| Rahimzadeh et al. (2021) | Federated capsule network learning | High | 91 | Medium | Yes |
| Ours (FLED-Block) | Federated Ensembled capsule networks | High | 98.5 | High | Yes |
These tables demonstrate that the FLED-Block model is well-suited for blockchain-based data sharing and retrieval, ensuring privacy and security while achieving superior diagnostic accuracy. Compared to the federated model proposed by He et al. (2020), FLED-Block exhibits a notable 7% increase in trust level, attributed to the integration of chaotic encryption and enhanced detection accuracy through the capsule network and ELM ensemble.
Furthermore, we compared the time and space complexity of the FLED-Block model with state-of-the-art classical models. Time complexity was assessed using Big-O notation. Classical models, operating on centralized systems, exhibit time complexity scaling as O(n2n), where n represents the number of computations. FLED-Block, leveraging a distributed system, reduces computational time based on the number of nodes used for training. In our experiments with 5 nodes, n was effectively reduced to five. Table 8 presents a comparative analysis of time and space complexity.
Table 8. Comparative Analysis Between the Blockchain-Based Model with Other Learning Models.
| References | Proposed model in blockchain | Time complexity | Space complexity |
|---|---|---|---|
| Parnian et al. (2020) | ResNETS | O(n2n) | 6.92 MB |
| He et al. (2020) | 2D-CNN | O(n2n-1) | 5.54 MB |
| Rahimzadeh et al. (2021) | Federated capsule network learning | O(n2n-5) | 3.25 MB |
| Ours (FLED-Block) | Federated ensembled capsule networks | O(n2n-5) | 2.85 MB |
Table 8 shows that the federated learning models achieve significantly lower time complexity compared to classical centralized models due to their distributed nature. Space complexity, measured by memory utilization, is also lower for federated models. The FLED-Block model, incorporating an extreme learning machine, further reduces space complexity due to the feedforward nature of ELMs, making it highly efficient in terms of both computational time and memory usage.
Conclusion: FLED-Block – A Promising Framework for Secure and Accurate AI in Healthcare
Traditional AI techniques often rely on centralized data storage and training, leading to computational bottlenecks and privacy concerns. To address these limitations, this paper introduces the blockchain-empowered FLED-Block framework, designed to enhance the analysis of heterogeneous CT images from multiple sources. FLED-Block facilitates secure data sharing among hospitals while maintaining stringent privacy and security protocols. The integration of an ensemble of capsule networks and extreme learning machines enables effective feature extraction and classification for accurate COVID-19 detection across diverse, publicly available CT image datasets. Federated learning, supported by blockchain technology, enables collaborative training among hospitals, ensuring data privacy and model robustness. The incorporation of chaotic encryption keys further enhances data retrieval and sharing security, bolstering trust in the system.
Extensive experimental evaluations and comparisons with other deep learning algorithms and blockchain-based federated models demonstrate the superior performance of FLED-Block. The model achieves high accuracy, precision, recall, and F1-score, proving to be more effective and trustworthy than existing approaches. While FLED-Block shows promising results, further improvements are needed to handle real-time clinical databases and reduce false positives. Future research will focus on reducing blockchain latency and optimizing the cost-effectiveness of the FLED-Block solution for practical clinical deployment and broader applications in secure and privacy-preserving healthcare AI.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
RD contributed to the conception and design of the study. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
[References list from the original article, maintaining original formatting and links]
Associated Data
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.